Dong Xu
Department of Radiology, Zhejiang Cancer Hospital, Hangzhou, 310022, China, Hangzhou Institute of Medicine, Key Laboratory of Head and Neck Cancer Translational Research of Zhejiang Province, Hangzhou, 310022, China, Zhejiang Provincial Research Center for Cancer Intelligent Diagnosis and Molecular Technology, Hangzhou, 310000, China, Wenling Medical Big Data and Artificial Intelligence Research Institute, 24th Floor, Machang Road, Taizhou, 310061, China, Taizhou Key Laboratory of Minimally Invasive Interventional Therapy and Artificial Intelligence, Taizhou Campus of Zhejiang Cancer Hospital
RIGA-Fold: A General Framework for Protein Inverse Folding via Recurrent Interaction and Geometric Awareness
Feb 04, 2026Abstract:Protein inverse folding, the task of predicting amino acid sequences for desired structures, is pivotal for de novo protein design. However, existing GNN-based methods typically suffer from restricted receptive fields that miss long-range dependencies and a "single-pass" inference paradigm that leads to error accumulation. To address these bottlenecks, we propose RIGA-Fold, a framework that synergizes Recurrent Interaction with Geometric Awareness. At the micro-level, we introduce a Geometric Attention Update (GAU) module where edge features explicitly serve as attention keys, ensuring strictly SE(3)-invariant local encoding. At the macro-level, we design an attention-based Global Context Bridge that acts as a soft gating mechanism to dynamically inject global topological information. Furthermore, to bridge the gap between structural and sequence modalities, we introduce an enhanced variant, RIGA-Fold*, which integrates trainable geometric features with frozen evolutionary priors from ESM-2 and ESM-IF via a dual-stream architecture. Finally, a biologically inspired ``predict-recycle-refine'' strategy is implemented to iteratively denoise sequence distributions. Extensive experiments on CATH 4.2, TS50, and TS500 benchmarks demonstrate that our geometric framework is highly competitive, while RIGA-Fold* significantly outperforms state-of-the-art baselines in both sequence recovery and structural consistency.
Unveiling Scaling Behaviors in Molecular Language Models: Effects of Model Size, Data, and Representation
Jan 30, 2026Abstract:Molecular generative models, often employing GPT-style language modeling on molecular string representations, have shown promising capabilities when scaled to large datasets and model sizes. However, it remains unclear and subject to debate whether these models adhere to predictable scaling laws under fixed computational budgets, which is a crucial understanding for optimally allocating resources between model size, data volume, and molecular representation. In this study, we systematically investigate the scaling behavior of molecular language models across both pretraining and downstream tasks. We train 300 models and conduct over 10,000 experiments, rigorously controlling compute budgets while independently varying model size, number of training tokens, and molecular representation. Our results demonstrate clear scaling laws in molecular models for both pretraining and downstream transfer, reveal the substantial impact of molecular representation on performance, and explain previously observed inconsistencies in scaling behavior for molecular generation. Additionally, we publicly release the largest library of molecular language models to date to facilitate future research and development. Code and models are available at https://github.com/SZU-ADDG/MLM-Scaling.
From Tokens to Blocks: A Block-Diffusion Perspective on Molecular Generation
Jan 29, 2026Abstract:Drug discovery can be viewed as a combinatorial search over an immense chemical space, motivating the development of deep generative models for de novo molecular design. Among these, GPT-based molecular language models (MLM) have shown strong molecular design performance by learning chemical syntax and semantics from large-scale data. However, existing MLMs face two fundamental limitations: they inadequately capture the graph-structured nature of molecules when formulated as next-token prediction problems, and they typically lack explicit mechanisms for target-aware generation. Here, we propose SoftMol, a unified framework that co-designs molecular representation, model architecture, and search strategy for target-aware molecular generation. SoftMol introduces soft fragments, a rule-free block representation of SMILES that enables diffusion-native modeling, and develops SoftBD, the first block-diffusion molecular language model that combines local bidirectional diffusion with autoregressive generation under molecular structural constraints. To favor generated molecules with high drug-likeness and synthetic accessibility, SoftBD is trained on a carefully curated dataset named ZINC-Curated. SoftMol further integrates a gated Monte Carlo tree search to assemble fragments in a target-aware manner. Experimental results show that, compared with current state-of-the-art models, SoftMol achieves 100% chemical validity, improves binding affinity by 9.7%, yields a 2-3x increase in molecular diversity, and delivers a 6.6x speedup in inference efficiency. Code is available at https://github.com/szu-aicourse/softmol
LoD-Structured 3D Gaussian Splatting for Streaming Video Reconstruction
Jan 26, 2026Abstract:Free-Viewpoint Video (FVV) reconstruction enables photorealistic and interactive 3D scene visualization; however, real-time streaming is often bottlenecked by sparse-view inputs, prohibitive training costs, and bandwidth constraints. While recent 3D Gaussian Splatting (3DGS) has advanced FVV due to its superior rendering speed, Streaming Free-Viewpoint Video (SFVV) introduces additional demands for rapid optimization, high-fidelity reconstruction under sparse constraints, and minimal storage footprints. To bridge this gap, we propose StreamLoD-GS, an LoD-based Gaussian Splatting framework designed specifically for SFVV. Our approach integrates three core innovations: 1) an Anchor- and Octree-based LoD-structured 3DGS with a hierarchical Gaussian dropout technique to ensure efficient and stable optimization while maintaining high-quality rendering; 2) a GMM-based motion partitioning mechanism that separates dynamic and static content, refining dynamic regions while preserving background stability; and 3) a quantized residual refinement framework that significantly reduces storage requirements without compromising visual fidelity. Extensive experiments demonstrate that StreamLoD-GS achieves competitive or state-of-the-art performance in terms of quality, efficiency, and storage.
Rethinking Drug-Drug Interaction Modeling as Generalizable Relation Learning
Jan 22, 2026Abstract:Drug-drug interaction (DDI) prediction is central to drug discovery and clinical development, particularly in the context of increasingly prevalent polypharmacy. Although existing computational methods achieve strong performance on standard benchmarks, they often fail to generalize to realistic deployment scenarios, where most candidate drug pairs involve previously unseen drugs and validated interactions are scarce. We demonstrate that proximity in the embedding spaces of prevailing molecule-centric DDI models does not reliably correspond to interaction labels, and that simply scaling up model capacity therefore fails to improve generalization. To address these limitations, we propose GenRel-DDI, a generalizable relation learning framework that reformulates DDI prediction as a relation-centric learning problem, in which interaction representations are learned independently of drug identities. This relation-level abstraction enables the capture of transferable interaction patterns that generalize to unseen drugs and novel drug pairs. Extensive experiments across multiple benchmark demonstrate that GenRel-DDI consistently and significantly outperforms state-of-the-art methods, with particularly large gains on strict entity-disjoint evaluations, highlighting the effectiveness and practical utility of relation learning for robust DDI prediction. The code is available at https://github.com/SZU-ADDG/GenRel-DDI.
Learning Diffusion Policy from Primitive Skills for Robot Manipulation
Jan 05, 2026Abstract:Diffusion policies (DP) have recently shown great promise for generating actions in robotic manipulation. However, existing approaches often rely on global instructions to produce short-term control signals, which can result in misalignment in action generation. We conjecture that the primitive skills, referred to as fine-grained, short-horizon manipulations, such as ``move up'' and ``open the gripper'', provide a more intuitive and effective interface for robot learning. To bridge this gap, we propose SDP, a skill-conditioned DP that integrates interpretable skill learning with conditional action planning. SDP abstracts eight reusable primitive skills across tasks and employs a vision-language model to extract discrete representations from visual observations and language instructions. Based on them, a lightweight router network is designed to assign a desired primitive skill for each state, which helps construct a single-skill policy to generate skill-aligned actions. By decomposing complex tasks into a sequence of primitive skills and selecting a single-skill policy, SDP ensures skill-consistent behavior across diverse tasks. Extensive experiments on two challenging simulation benchmarks and real-world robot deployments demonstrate that SDP consistently outperforms SOTA methods, providing a new paradigm for skill-based robot learning with diffusion policies.
Diagnostic Performance of Universal-Learning Ultrasound AI Across Multiple Organs and Tasks: the UUSIC25 Challenge
Dec 19, 2025Abstract:IMPORTANCE: Current ultrasound AI remains fragmented into single-task tools, limiting clinical utility compared to versatile modern ultrasound systems. OBJECTIVE: To evaluate the diagnostic accuracy and efficiency of single general-purpose deep learning models for multi-organ classification and segmentation. DESIGN: The Universal UltraSound Image Challenge 2025 (UUSIC25) involved developing algorithms on 11,644 images (public/private). Evaluation used an independent, multi-center test set of 2,479 images, including data from a center completely unseen during training to assess generalization. OUTCOMES: Diagnostic performance (Dice Similarity Coefficient [DSC]; Area Under the Receiver Operating Characteristic Curve [AUC]) and computational efficiency (inference time, GPU memory). RESULTS: Of 15 valid algorithms, the top model (SMART) achieved a macro-averaged DSC of 0.854 across 5 segmentation tasks and AUC of 0.766 for binary classification. Models showed high capability in segmentation (e.g., fetal head DSC: 0.942) but variability in complex tasks subject to domain shift. Notably, in breast cancer molecular subtyping, the top model's performance dropped from AUC 0.571 (internal) to 0.508 (unseen external center), highlighting generalization challenges. CONCLUSIONS: General-purpose AI models achieve high accuracy and efficiency across multiple tasks using a single architecture. However, performance degradation on unseen data suggests domain generalization is critical for future clinical deployment.
Toward Closed-loop Molecular Discovery via Language Model, Property Alignment and Strategic Search
Dec 18, 2025Abstract:Drug discovery is a time-consuming and expensive process, with traditional high-throughput and docking-based virtual screening hampered by low success rates and limited scalability. Recent advances in generative modelling, including autoregressive, diffusion, and flow-based approaches, have enabled de novo ligand design beyond the limits of enumerative screening. Yet these models often suffer from inadequate generalization, limited interpretability, and an overemphasis on binding affinity at the expense of key pharmacological properties, thereby restricting their translational utility. Here we present Trio, a molecular generation framework integrating fragment-based molecular language modeling, reinforcement learning, and Monte Carlo tree search, for effective and interpretable closed-loop targeted molecular design. Through the three key components, Trio enables context-aware fragment assembly, enforces physicochemical and synthetic feasibility, and guides a balanced search between the exploration of novel chemotypes and the exploitation of promising intermediates within protein binding pockets. Experimental results show that Trio reliably achieves chemically valid and pharmacologically enhanced ligands, outperforming state-of-the-art approaches with improved binding affinity (+7.85%), drug-likeness (+11.10%) and synthetic accessibility (+12.05%), while expanding molecular diversity more than fourfold. By combining generalization, plausibility, and interpretability, Trio establishes a closed-loop generative paradigm that redefines how chemical space can be navigated, offering a transformative foundation for the next era of AI-driven drug discovery.
4DGS-Craft: Consistent and Interactive 4D Gaussian Splatting Editing
Oct 02, 2025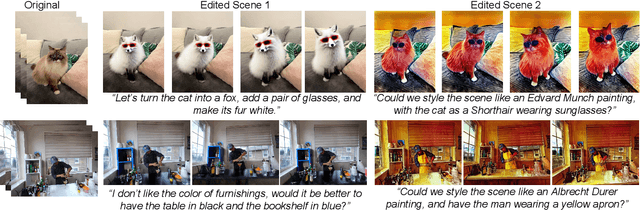
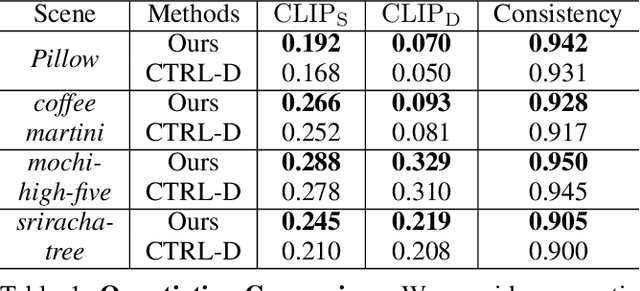
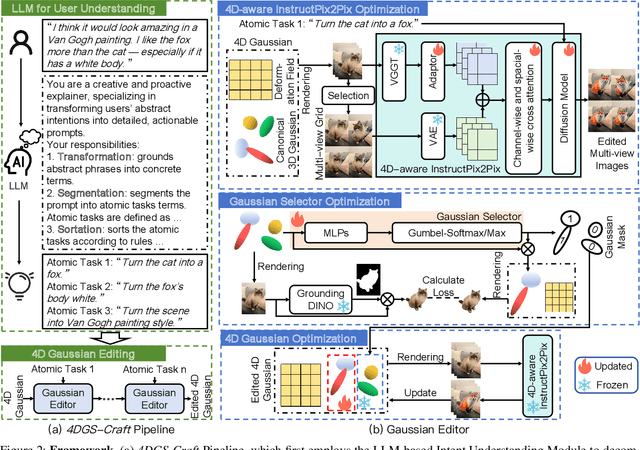
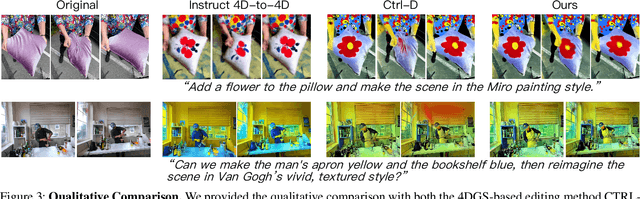
Abstract:Recent advances in 4D Gaussian Splatting (4DGS) editing still face challenges with view, temporal, and non-editing region consistency, as well as with handling complex text instructions. To address these issues, we propose 4DGS-Craft, a consistent and interactive 4DGS editing framework. We first introduce a 4D-aware InstructPix2Pix model to ensure both view and temporal consistency. This model incorporates 4D VGGT geometry features extracted from the initial scene, enabling it to capture underlying 4D geometric structures during editing. We further enhance this model with a multi-view grid module that enforces consistency by iteratively refining multi-view input images while jointly optimizing the underlying 4D scene. Furthermore, we preserve the consistency of non-edited regions through a novel Gaussian selection mechanism, which identifies and optimizes only the Gaussians within the edited regions. Beyond consistency, facilitating user interaction is also crucial for effective 4DGS editing. Therefore, we design an LLM-based module for user intent understanding. This module employs a user instruction template to define atomic editing operations and leverages an LLM for reasoning. As a result, our framework can interpret user intent and decompose complex instructions into a logical sequence of atomic operations, enabling it to handle intricate user commands and further enhance editing performance. Compared to related works, our approach enables more consistent and controllable 4D scene editing. Our code will be made available upon acceptance.
TOM: An Open-Source Tongue Segmentation Method with Multi-Teacher Distillation and Task-Specific Data Augmentation
Aug 19, 2025Abstract:Tongue imaging serves as a valuable diagnostic tool, particularly in Traditional Chinese Medicine (TCM). The quality of tongue surface segmentation significantly affects the accuracy of tongue image classification and subsequent diagnosis in intelligent tongue diagnosis systems. However, existing research on tongue image segmentation faces notable limitations, and there is a lack of robust and user-friendly segmentation tools. This paper proposes a tongue image segmentation model (TOM) based on multi-teacher knowledge distillation. By incorporating a novel diffusion-based data augmentation method, we enhanced the generalization ability of the segmentation model while reducing its parameter size. Notably, after reducing the parameter count by 96.6% compared to the teacher models, the student model still achieves an impressive segmentation performance of 95.22% mIoU. Furthermore, we packaged and deployed the trained model as both an online and offline segmentation tool (available at https://itongue.cn/), allowing TCM practitioners and researchers to use it without any programming experience. We also present a case study on TCM constitution classification using segmented tongue patches. Experimental results demonstrate that training with tongue patches yields higher classification performance and better interpretability than original tongue images. To our knowledge, this is the first open-source and freely available tongue image segmentation tool.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge